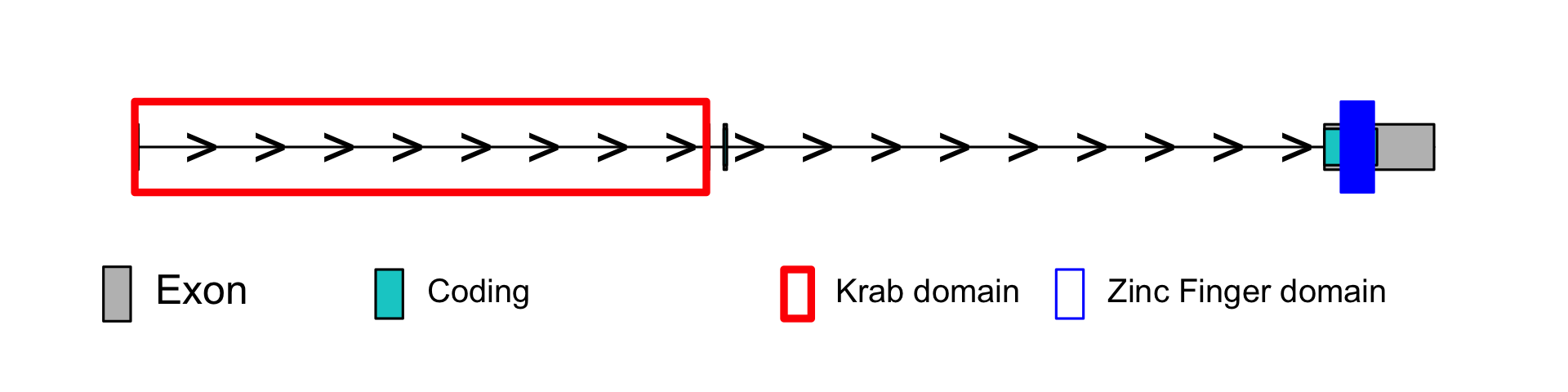
<!-- bara: put body text here -->
<u>Section list:</u>
<ul>
<li><a href='#General'>General</a></li>
<li><a href='#TE enrichment'>TE enrichment</a></li>
<li><a href='#Age'>Cross-Species Sequence Comparison</a></li>
<li><a href='#Polymorphism'>Polymorphism</a></li>
<li><a href='#Expression in tissues (from GTEx)'>Expression in tissues (from GTEx)</a></li>
<li><a href='#CisBP motifs'>CisBP motifs</a></li>
<li><a href='#Genomic regions'>Genomic regions</a></li>
<li><a href='#Genomic Location'>Genomic Location</a></li>
<li><a href='#MSA'>MSA of most enriched TE subfamilies</a></li>
<li><a href='#ZFP'>Zinc Finger Print alignment</a></li>
</ul>
------
<h2 id='General'>General</h2>
| | |
|--|--|
| <b>Gene ensembl <br><i>Link to the corresponding Ensembl site</i></b> | <a href="http://www.ensembl.org/Multi/Search/Results?q=ENSG00000213096;site=ensembl~">ENSG00000213096</a> |
| <b>Age (years) <br><i>Estimated age of the KRAB-ZNF as in <a href="https://pubmed.ncbi.nlm.nih.gov/28273063/">Imbeault et al</a></i></b> | 43.1 |
| <b>Age (species) <br><i>The name of the species where the KRAB-ZNF was first observed as in <a href="https://pubmed.ncbi.nlm.nih.gov/28273063/">Imbeault et al</a></i></b> | Nancy Ma's night monkey |
| <b>\#peaks in exo <br><i>Number of peaks found in the ChIP-exo dataset</i></b> | 949 |
| <b>Incomplete KRAB domain</b> | Yes |
| <b>Interacts with K1</b> | Not in database |
| <b>Interacts with</b> | Not in database |
| <b>ORF sequence</b> | <a href="../ORFseq/ZNF254.txt">Open</a> |
| <b>Zinc fingers</b> | 13 |
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id='TE enrichment'>TE enrichment</h2>
<table>
<tr><th>Family</th><th>Subfamily</th></tr>
<tr><td><table></table>
<input type="button" id="button_TE1" style="float:left" value="[+] Help" onclick="afficher_TE1()" /><br>
<form id="doc_TE1" style="display: none;">
<br>Enrichment of peaks over different repetitive element families, calculated with pyTEnrich v.0.6. The number of peaks over each family is shown by the colored bars and indicated by the numbers above them, the expected number of peaks following a random distribution is shown as black transparent bars. The adj. p-value of the enrichment is shown on the right (adj. p < 0.0001 = ****, < 0.001=***, < 0.01=**, < 0.05=*, >= 0.05 = n.s). Rows are ordered by adj. p-value. The number next to the title indicates the total number of peaks for the experiment.
<br><img src="../TEbound/legend_for_bara.png" width="200" height="200"/></form><br>
<a href="../TEbound/fam_viz/ZNF254_fam.pdf">
<img src="../TEbound/fam_png/ZNF254_fam.png" width="400" height="400" /></a>
</td><td><table></table>
<input type="button" id="button_TE2" style="float:left" value="[+] Help" onclick="afficher_TE2()" /><br>
<form id="doc_TE2" style="display: none;">
<br>Enrichment of peaks over different repetitive element subfamilies, calculated with pyTEnrich v.0.6. Subfamilies with adj. p < 0.01 are shown. The width of the colored bars represents the number of peaks per subfamily also shown as a number above the bar, whereas the black transparent bars represents the expected number of peaks following a random distribution. The adj. p-value of the enrichment is shown with stars (adj. p < 0.0001 = ****, < 0.001=***, < 0.01=**, < 0.05=*, >= 0.05 = n.s). Rows are ordered by adj. p-value. The number next to the title indicates the total number of peaks for the experiment.
<br><img src="../TEbound/legend_for_bara.png" width="200" height="200"/></form><br>
<a href="../TEbound/sfam_viz/ZNF254_subfam.pdf">
<img src="../TEbound/sfam_png/ZNF254_subfam.png" width="400" height="400" /></a>
</td></tr>
</table>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id='Age'>Cross-Species Sequence Comparison</h2>
<input type="button" id="button_age" style="float:left" value="[+] Help" onclick="afficher_age()" /><br>
<form id="doc_age" style="display: none;">
The 4 amino acids known to bind DNA were extracted from Zinc Finger protein sequence (positions -1, 2, 3, 6). Based on this subset of amino acids, we aligned the human Zinc Finger to every other Zinc Fingers in human and 68 foreign species. In the plot X.X, the alignment scores between the Zinc Finger of interest and other Zinc Fingers was drawn as one boxplot for each species. The yellow line joins the dots corresponding to the best alignment scores found for each species. Same was done for the KRAB domain, but only showing the best alignments joined by a red line. A dashed vertical red line shows the age estimate from Imbeault et al. 2018.
</form><br>
<a href='../EvolLine/EvoLine_ZNF254.pdf'>
<img src="../EvolLine/EvolLine/EvoLine_ZNF254.png" height="400" width="400" />
</a>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id='Polymorphism'>Polymorphism</h2>
<table>
<tr><th>gnomAD genome</th><th>gnomAD exome</th></tr>
<tr><td><table></table>
| Region | #miss.SNP x Kb |
|--|--|
| Exon | 78.901 |
| Exon (no domain, nor Zfinger) | 78.901 |
| Krab domain | NA |
| Zinc finger | 91.416 |
| Region | #miss.SNP |
|--|--|
| Zinc finger | 54 |
| C2H2 | 14 |
| Zinc finger print | 10 |
<input type="button" id="button" style="float:left" value="[+] Help" onclick="afficher()" /><br>
<form id="doc" style="display: none;">
<br>Gene view with all variants found at the gene of interest (canonical transcript).<br><ul><li> blue dots: synonymous & noncoding variants.</li><li> red dots: missense variants.</li><li> orange dots: splice variants.</li><li> green dots: Loss of function variants (frameshift, stop_lost or stop_gained mutation).</li><li> GERP score: a conservation score widely used to look at constraint across genes and non-coding regions, and also used in the best pathogenic variant scoring softwares. A score above 2 is considered conserved.</li><li> Green boxes on the genes: the exons.</li><li> Red boxes: the KRAB domain.</li><li> Blue boxes: the zinc finger domains.</li><li> Pink boxes: degenerate zinc finger domains.</li></ul>
</form><br>
<a href='../MAF_gnomAD/ZNF254.pdf'>
<img src="../MAF_gnomAD/MAF_gnomAD/ZNF254.png" width="400" height="200" />
</a>
<input type="button" id="button_2" style="float:left" value="[+] Help" onclick="afficher_2()" /><br>
<form id="doc_2" style="display: none;">
<br>Zoomed plot of variants in each zinc finger domain with the specific amino acids plotted.<ul><li>blue dots: synonymous & noncoding variants.</li><li>red dots: missense variants.</li><li>orange dots: splice variants.</li><li>green dots: Loss of function variants (frameshift, stop_lost or stop_gained mutation).</li><li> Blue boxes: The positions of the DNA contacting amino acids</li><li> Red boxes: The Cytidine and Histidine amino acid positions</li></ul>
</form><br>
<a href='../Zfinger_gnomAD/ZNF254.pdf'>
<img src="../Zfinger_gnomAD/Zfinger_gnomAD/ZNF254.png" width="400" height="200" />
</a>
</td><td><table></table>
| Region | #miss.SNP x Kb |
|--|--|
| Exon | 161.835 |
| Exon (no domain, nor Zfinger) | 161.835 |
| Krab domain | NA |
| Zinc finger | 360.089 |
| Region | #miss.SNP |
|--|--|
| Zinc finger | 231 |
| C2H2 | 45 |
| Zinc finger print | 34 |
<input type="button" id="button1" style="float:left" value="[+] Help" onclick="afficher1()" /><br>
<form id="doc1" style="display: none;">
<br>Gene view with all variants found at the gene of interest (canonical transcript).<br><ul><li> blue dots: synonymous & noncoding variants.</li><li> red dots: missense variants.</li><li> orange dots: splice variants.</li><li> green dots: Loss of function variants (frameshift, stop_lost or stop_gained mutation).</li><li> GERP score: a conservation score widely used to look at constraint across genes and non-coding regions, and also used in the best pathogenic variant scoring softwares. A score above 2 is considered conserved.</li><li> Green boxes on the genes: the exons.</li><li> Red boxes: the KRAB domain.</li><li> Blue boxes: the zinc finger domains.</li><li> Pink boxes: degenerate zinc finger domains.</li></ul>
</form><br>
<a href='../MAF_gnomAD_exome/ZNF254.pdf'>
<img src="../MAF_gnomAD_exome/MAF_gnomAD_exome/ZNF254.png" width="400" height="200" />
</a>
<input type="button" id="button2" style="float:left" value="[+] Help" onclick="afficher2()" /><br>
<form id="doc2" style="display: none;">
<br>Zoomed plot of variants in each zinc finger domain with the specific amino acids plotted.<ul><li>blue dots: synonymous & noncoding variants.</li><li>red dots: missense variants.</li><li>orange dots: splice variants.</li><li>green dots: Loss of function variants (frameshift, stop_lost or stop_gained mutation).</li><li> Blue boxes: The positions of the DNA contacting amino acids</li><li> Red boxes: The Cytidine and Histidine amino acid positions</li></ul>
</form><br>
<a href='../Zfinger_gnomAD_exome/ZNF254.pdf'>
<img src="../Zfinger_gnomAD_exome/Zfinger_gnomAD_exome/ZNF254.png" width="400" height="200" />
</a>
</td></tr>
</table>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id='Expression in tissues (from GTEx)'>Expression in tissues (from GTEx)</h2>
<input type="button" id="GTX_button" style="float:left" value="Open Expression Information from GTEx Website" onclick="document.location.href='https://www.gtexportal.org/home/gene/ZNF254'" /><br><br>
<a href="../GTEx_cecile/Figures_transcripts/Histogram_coding/Histogram_ZNF254.png"><img src="../GTEx_cecile/Figures_transcripts/Histogram_coding/Histogram_ZNF254.png" height="400" width="400" /></a>
<a href="../GTEx_cecile/Figures_transcripts/Heatmap_coding/Heatmap_C_ZNF254.png"><img src="../GTEx_cecile/Figures_transcripts/Heatmap_coding/Heatmap_C_ZNF254.png" height="400" width="400" /></a><br>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id='CisBP motifs'>CisBP motifs</h2>
<input type="button" id="CisBP_button" style="float:left" value="Open CisBP Website" onclick="document.location.href='http://cisbp.ccbr.utoronto.ca'" /><br><br>
<table>
<tr><th>Forward</th><th>Reverse</th></tr>
<tr><td>
<tr><td><table><tr><th> ENSEMBL ID </th><td> ENSG00000213096 </td></tr><tr><th> Motif_ID </th><td> M07760_2.00 </td></tr><tr><th> Motif_Type </th><td> ChIP-seq+ChIP-exo </td></tr><tr><th> MSource_Identifier </th><td> Barazandeh2018 </td></tr><tr><th> Study_ID </th><td> ZNF254.RCADE.OriginalMapping </td></tr><tr><th> Author_Year </th><td> Barazandeh_2018 </td></tr><tr><th> PMID </th><td> 29146583 </td></tr><tr><th> TF_Status </th><td> D </td></tr></table><a href="../logos_kzfp/M07760_2.00_fwd.png"><img src="../logos_kzfp/M07760_2.00_fwd.png" width="200" height="100"></td>
<td><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><a href="../logos_kzfp/M07760_2.00_rev.png"><img src="../logos_kzfp/M07760_2.00_rev.png" width="200" height="100"></td></tr>
</td></tr>
</table>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id='Genomic regions'>Genomic regions</h2>
<a href='../figs/ZNF254.pdf'>
<img src="../figs/figs/ZNF254.png" width="400" height="400" /><br><br>
</a>
<input type="button" id="GenReg_button" style="float:left" value="Open Table with Annotation for Each Bound Loci" onclick="document.location.href='../tables/ZNF254_overlaps.xls'" /><br><br>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id='Genomic Location'>Genomic Location</h2>
<input type="button" id="button_gen_loc" style="float:left" value="[+] Help" onclick="afficher_gen_loc()" /><br>
<form id="doc_gen_loc" style="display: none;">
<br>This map shows the position (dot) of each pair of a KRAB and Zinc Finger domains (KZFs) in the human genome, including non protein coding ones, as well as the different clusters of KZFs (light blue regions). <br>The dots are colored according to the age of the KZFs [Reference to ACs work. Probably Christians Paper] and dark grey if no age could be assigned. <br>The chromosomes are shown as grey bars with the black regions depicting the centromeres.
</form><br>
<a href='../make_static_plots/KRABmap_ZNF254.pdf'>
<img src="../make_static_plots/make_static_plots/KRABmap_ZNF254.png" width="800" height="430" />
</a>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id="MSA">MSA of most enriched TE subfamilies</h2>
<input type="button" id="button_msa" style="float:left" value="[+] Help" onclick="afficher_msa()"/><br>
<form id="doc_msa" style="display: none;">
<br>Multiple Sequence Alignment of most enriched TE subfamilies.
</form>
<a href="../MSA_pdfs/ZNF254.pdf">
<img src="../MSA_pdfs/pngs/plotmsa_ZNF254.png" width="800" height="430" />
</a>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id="ZFP">ZFP alignment</h2>
<input type="button" id="button_zfp" style="float:left" value="[+] Help" onclick="afficher_zfp()"/><br>
<form id="doc_zfp" style="display: none;">
<br>Zinc Finger Print alignment of the ZFP of interest against all other ZFPs
</form>
<a href="../ZFP_CAZF_score/ZNF254.pdf">
<img src="../ZFP_CAZF_score/pngs/plotzfp_ZNF254.png" width="400" height="200" />
</a>
<table>
<tr>
<td></td>
<td>GeneName1</td>
<td>KZFP1</td>
<td>GeneName2</td>
<td>KZFP2</td>
<td>CAZF_score</td>
</tr>
<tr>
<td>956661</td>
<td>ZNF254</td>
<td>NC_000019.9_FF_OOFZFA_88</td>
<td>ZNF431</td>
<td>NC_000019.9_FF_OOFZFA_65</td>
<td>0.43496105164909</td>
</tr>
<tr>
<td>956659</td>
<td>ZNF254</td>
<td>NC_000019.9_FF_OOFZFA_88</td>
<td>ZNF430</td>
<td>NC_000019.9_FF_OOFZFA_63</td>
<td>0.360755808975874</td>
</tr>
<tr>
<td>956690</td>
<td>ZNF254</td>
<td>NC_000019.9_FF_OOFZFA_88</td>
<td>ZNF726</td>
<td>NC_000019.9_FF_OOFZFA_86</td>
<td>0.353789305897696</td>
</tr>
<tr>
<td>956684</td>
<td>ZNF254</td>
<td>NC_000019.9_FF_OOFZFA_88</td>
<td>ZNF91</td>
<td>NC_000019.9_RF_OOFZFA_378</td>
<td>0.326970917816965</td>
</tr>
<tr>
<td>956685</td>
<td>ZNF254</td>
<td>NC_000019.9_FF_OOFZFA_88</td>
<td>ZNF725P</td>
<td>NC_000019.9_RF_OOFZFA_380</td>
<td>0.3004329004329</td>
</tr>
<tr>
<td>956668</td>
<td>ZNF254</td>
<td>NC_000019.9_FF_OOFZFA_88</td>
<td>ZNF100</td>
<td>NC_000019.9_RF_OOFZFA_359</td>
<td>0.293678416594702</td>
</tr>
<tr>
<td>957066</td>
<td>ZNF254</td>
<td>NC_000019.9_FF_OOFZFA_88</td>
<td>ZNF100</td>
<td>NW_003315964.2_RF_OOFZFA_1</td>
<td>0.293678416594702</td>
</tr>
<tr>
<td>956660</td>
<td>ZNF254</td>
<td>NC_000019.9_FF_OOFZFA_88</td>
<td>ZNF714</td>
<td>NC_000019.9_FF_OOFZFA_64</td>
<td>0.290503231884437</td>
</tr>
<tr>
<td>956681</td>
<td>ZNF254</td>
<td>NC_000019.9_FF_OOFZFA_88</td>
<td>ZNF730</td>
<td>NC_000019.9_FF_OOFZFA_83</td>
<td>0.269864531267719</td>
</tr>
<tr>
<td>956665</td>
<td>ZNF254</td>
<td>NC_000019.9_FF_OOFZFA_88</td>
<td>ZNF493</td>
<td>NC_000019.9_FF_OOFZFA_68</td>
<td>0.25341140359657</td>
</tr>
</table>
<input type="button" id="button_csv" style="float:left" value="Open full table" onclick="document.location.href='../ZFP_CAZF_score/csv/ZNF254.xlsx'" /><br>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
<!-- bara: end of place to put text -->