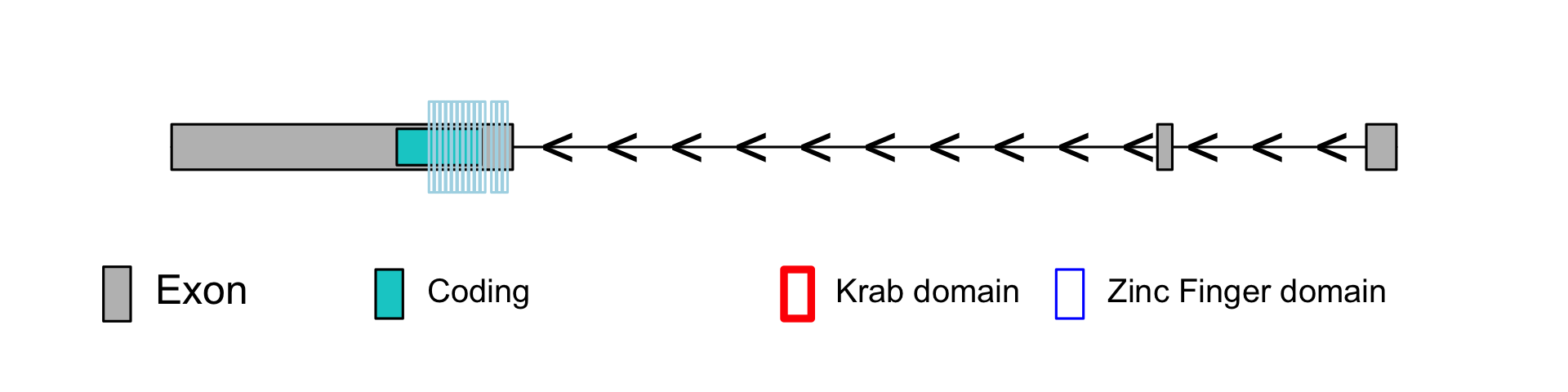
<!-- bara: put body text here -->
<u>Section list:</u>
<ul>
<li><a href='#General'>General</a></li>
<li><a href='#Polymorphism'>Polymorphism</a></li>
<li><a href='#Expression in tissues (from GTEx)'>Expression in tissues (from GTEx)</a></li>
<li><a href='#CisBP motifs'>CisBP motifs</a></li>
<li><a href='#ZFP'>Zinc Finger Print alignment</a></li>
</ul>
------
<h2 id='General'>General</h2>
| | |
|--|--|
| <b>Gene ensembl <br><i>Link to the corresponding Ensembl site</i></b> | <a href="http://www.ensembl.org/Multi/Search/Results?q=ENSG00000254004;site=ensembl~">ENSG00000254004</a> |
| <b>Age (years) <br><i>Estimated age of the KRAB-ZNF as in <a href="https://pubmed.ncbi.nlm.nih.gov/28273063/">Imbeault et al</a></i></b> | 97.5 |
| <b>Age (species) <br><i>The name of the species where the KRAB-ZNF was first observed as in <a href="https://pubmed.ncbi.nlm.nih.gov/28273063/">Imbeault et al</a></i></b> | Giant Panda |
| <b>\#peaks in exo <br><i>Number of peaks found in the ChIP-exo dataset</i></b> | |
| <b>Incomplete KRAB domain</b> | |
| <b>Interacts with K1</b> | Not in database |
| <b>Interacts with</b> | Not in database |
| <b>ORF sequence</b> | |
| <b>Zinc fingers</b> | 13 |
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id='Polymorphism'>Polymorphism</h2>
<table>
<tr><th>gnomAD genome</th><th>gnomAD exome</th></tr>
<tr><td><table></table>
| Region | #miss.SNP x Kb |
|--|--|
| Exon | 55.595 |
| Exon (no domain, nor Zfinger) | 55.595 |
| Krab domain | NA |
| Zinc finger | 54.627 |
| Region | #miss.SNP |
|--|--|
| Zinc finger | 24 |
| C2H2 | 0 |
| Zinc finger print | 0 |
<input type="button" id="button" style="float:left" value="[+] Help" onclick="afficher()" /><br>
<form id="doc" style="display: none;">
<br>Gene view with all variants found at the gene of interest (canonical transcript).<br><ul><li> blue dots: synonymous & noncoding variants.</li><li> red dots: missense variants.</li><li> orange dots: splice variants.</li><li> green dots: Loss of function variants (frameshift, stop_lost or stop_gained mutation).</li><li> GERP score: a conservation score widely used to look at constraint across genes and non-coding regions, and also used in the best pathogenic variant scoring softwares. A score above 2 is considered conserved.</li><li> Green boxes on the genes: the exons.</li><li> Red boxes: the KRAB domain.</li><li> Blue boxes: the zinc finger domains.</li><li> Pink boxes: degenerate zinc finger domains.</li></ul>
</form><br>
<a href='../MAF_gnomAD/ZNF260.pdf'>
<img src="../MAF_gnomAD/MAF_gnomAD/ZNF260.png" width="400" height="200" />
</a>
<input type="button" id="button_2" style="float:left" value="[+] Help" onclick="afficher_2()" /><br>
<form id="doc_2" style="display: none;">
<br>Zoomed plot of variants in each zinc finger domain with the specific amino acids plotted.<ul><li>blue dots: synonymous & noncoding variants.</li><li>red dots: missense variants.</li><li>orange dots: splice variants.</li><li>green dots: Loss of function variants (frameshift, stop_lost or stop_gained mutation).</li><li> Blue boxes: The positions of the DNA contacting amino acids</li><li> Red boxes: The Cytidine and Histidine amino acid positions</li></ul>
</form><br>
<a href='../Zfinger_gnomAD/ZNF260.pdf'>
<img src="../Zfinger_gnomAD/Zfinger_gnomAD/ZNF260.png" width="400" height="200" />
</a>
</td><td><table></table>
| Region | #miss.SNP x Kb |
|--|--|
| Exon | 46.241 |
| Exon (no domain, nor Zfinger) | 46.241 |
| Krab domain | NA |
| Zinc finger | 150.502 |
| Region | #miss.SNP |
|--|--|
| Zinc finger | 87 |
| C2H2 | 0 |
| Zinc finger print | 0 |
<input type="button" id="button1" style="float:left" value="[+] Help" onclick="afficher1()" /><br>
<form id="doc1" style="display: none;">
<br>Gene view with all variants found at the gene of interest (canonical transcript).<br><ul><li> blue dots: synonymous & noncoding variants.</li><li> red dots: missense variants.</li><li> orange dots: splice variants.</li><li> green dots: Loss of function variants (frameshift, stop_lost or stop_gained mutation).</li><li> GERP score: a conservation score widely used to look at constraint across genes and non-coding regions, and also used in the best pathogenic variant scoring softwares. A score above 2 is considered conserved.</li><li> Green boxes on the genes: the exons.</li><li> Red boxes: the KRAB domain.</li><li> Blue boxes: the zinc finger domains.</li><li> Pink boxes: degenerate zinc finger domains.</li></ul>
</form><br>
<a href='../MAF_gnomAD_exome/ZNF260.pdf'>
<img src="../MAF_gnomAD_exome/MAF_gnomAD_exome/ZNF260.png" width="400" height="200" />
</a>
<input type="button" id="button2" style="float:left" value="[+] Help" onclick="afficher2()" /><br>
<form id="doc2" style="display: none;">
<br>Zoomed plot of variants in each zinc finger domain with the specific amino acids plotted.<ul><li>blue dots: synonymous & noncoding variants.</li><li>red dots: missense variants.</li><li>orange dots: splice variants.</li><li>green dots: Loss of function variants (frameshift, stop_lost or stop_gained mutation).</li><li> Blue boxes: The positions of the DNA contacting amino acids</li><li> Red boxes: The Cytidine and Histidine amino acid positions</li></ul>
</form><br>
<a href='../Zfinger_gnomAD_exome/ZNF260.pdf'>
<img src="../Zfinger_gnomAD_exome/Zfinger_gnomAD_exome/ZNF260.png" width="400" height="200" />
</a>
</td></tr>
</table>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id='Expression in tissues (from GTEx)'>Expression in tissues (from GTEx)</h2>
<input type="button" id="GTX_button" style="float:left" value="Open Expression Information from GTEx Website" onclick="document.location.href='https://www.gtexportal.org/home/gene/ZNF260'" /><br><br>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id='CisBP motifs'>CisBP motifs</h2>
<input type="button" id="CisBP_button" style="float:left" value="Open CisBP Website" onclick="document.location.href='http://cisbp.ccbr.utoronto.ca'" /><br><br>
<table>
<tr><th>Forward</th><th>Reverse</th></tr>
<tr><td>
<tr><td><table><tr><th> ENSEMBL ID </th><td> ENSG00000254004 </td></tr><tr><th> Motif_ID </th><td> M08395_2.00 </td></tr><tr><th> Motif_Type </th><td> ChIP-seq </td></tr><tr><th> MSource_Identifier </th><td> Schmitges2016 </td></tr><tr><th> Study_ID </th><td> Q3ZCT1_9-13.RCADE </td></tr><tr><th> Author_Year </th><td> Schmitges_2016 </td></tr><tr><th> PMID </th><td> 27852650 </td></tr><tr><th> TF_Status </th><td> D </td></tr></table><a href="../logos_kzfp/M08395_2.00_fwd.png"><img src="../logos_kzfp/M08395_2.00_fwd.png" width="200" height="100"></td>
<td><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><a href="../logos_kzfp/M08395_2.00_rev.png"><img src="../logos_kzfp/M08395_2.00_rev.png" width="200" height="100"></td></tr>
<tr><td><table><tr><th> ENSEMBL ID </th><td> ENSG00000254004 </td></tr><tr><th> Motif_ID </th><td> M08959_2.00 </td></tr><tr><th> Motif_Type </th><td> Misc </td></tr><tr><th> MSource_Identifier </th><td> HocoMoco </td></tr><tr><th> Study_ID </th><td> ZN260_HUMAN.H11MO.0.C </td></tr><tr><th> Author_Year </th><td> Kulakovskiy_2013 </td></tr><tr><th> PMID </th><td> 23175603 </td></tr><tr><th> TF_Status </th><td> D </td></tr></table><a href="../logos_kzfp/M08959_2.00_fwd.png"><img src="../logos_kzfp/M08959_2.00_fwd.png" width="200" height="100"></td>
<td><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><a href="../logos_kzfp/M08959_2.00_rev.png"><img src="../logos_kzfp/M08959_2.00_rev.png" width="200" height="100"></td></tr>
</td></tr>
</table>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
------
<h2 id="ZFP">ZFP alignment</h2>
<input type="button" id="button_zfp" style="float:left" value="[+] Help" onclick="afficher_zfp()"/><br>
<form id="doc_zfp" style="display: none;">
<br>Zinc Finger Print alignment of the ZFP of interest against all other ZFPs
</form>
<a href="../ZFP_CAZF_score/ZNF260.pdf">
<img src="../ZFP_CAZF_score/pngs/plotzfp_ZNF260.png" width="400" height="200" />
</a>
<table>
<tr>
<td></td>
<td>GeneName1</td>
<td>KZFP1</td>
<td>GeneName2</td>
<td>KZFP2</td>
<td>CAZF_score</td>
</tr>
<tr>
<td>984552</td>
<td>ZNF260</td>
<td>NC_000019.9_RF_OOFZFA_410</td>
<td>ZNF568</td>
<td>NC_000019.9_FF_OOFZFA_127</td>
<td>0.170096403411198</td>
</tr>
<tr>
<td>984561</td>
<td>ZNF260</td>
<td>NC_000019.9_RF_OOFZFA_410</td>
<td>ZNF569</td>
<td>NC_000019.9_RF_OOFZFA_420</td>
<td>0.147910709979675</td>
</tr>
<tr>
<td>983786</td>
<td>ZNF260</td>
<td>NC_000019.9_RF_OOFZFA_410</td>
<td>ZNF678</td>
<td>NC_000001.10_FF_OOFZFA_479</td>
<td>0.134467634603751</td>
</tr>
<tr>
<td>984644</td>
<td>ZNF260</td>
<td>NC_000019.9_RF_OOFZFA_410</td>
<td>ZNF677</td>
<td>NC_000019.9_RF_OOFZFA_489</td>
<td>0.126431556948798</td>
</tr>
<tr>
<td>984443</td>
<td>ZNF260</td>
<td>NC_000019.9_RF_OOFZFA_410</td>
<td>ZNF554</td>
<td>NC_000019.9_FF_OOFZFA_2</td>
<td>0.115864050733254</td>
</tr>
<tr>
<td>984108</td>
<td>ZNF260</td>
<td>NC_000019.9_RF_OOFZFA_410</td>
<td>ZNF16</td>
<td>NC_000008.10_RF_OOFZFA_781</td>
<td>0.112791991101224</td>
</tr>
<tr>
<td>983805</td>
<td>ZNF260</td>
<td>NC_000019.9_RF_OOFZFA_410</td>
<td>ZNF514</td>
<td>NC_000002.11_RF_OOFZFA_821</td>
<td>0.112772493063813</td>
</tr>
<tr>
<td>984029</td>
<td>ZNF260</td>
<td>NC_000019.9_RF_OOFZFA_410</td>
<td>ZNF138</td>
<td>NC_000007.13_FF_OOFZFA_175</td>
<td>0.111760710553814</td>
</tr>
<tr>
<td>984035</td>
<td>ZNF260</td>
<td>NC_000019.9_RF_OOFZFA_410</td>
<td>ZNF394</td>
<td>NC_000007.13_RF_OOFZFA_632</td>
<td>0.111567309565556</td>
</tr>
<tr>
<td>984719</td>
<td>ZNF260</td>
<td>NC_000019.9_RF_OOFZFA_410</td>
<td>ZNF8</td>
<td>NC_000019.9_FF_OOFZFA_262</td>
<td>0.111226714229092</td>
</tr>
</table>
<input type="button" id="button_csv" style="float:left" value="Open full table" onclick="document.location.href='../ZFP_CAZF_score/csv/ZNF260.xlsx'" /><br>
<div class="adroite"><a href="#"><b>back to top</b></a></div>
<!-- bara: end of place to put text -->